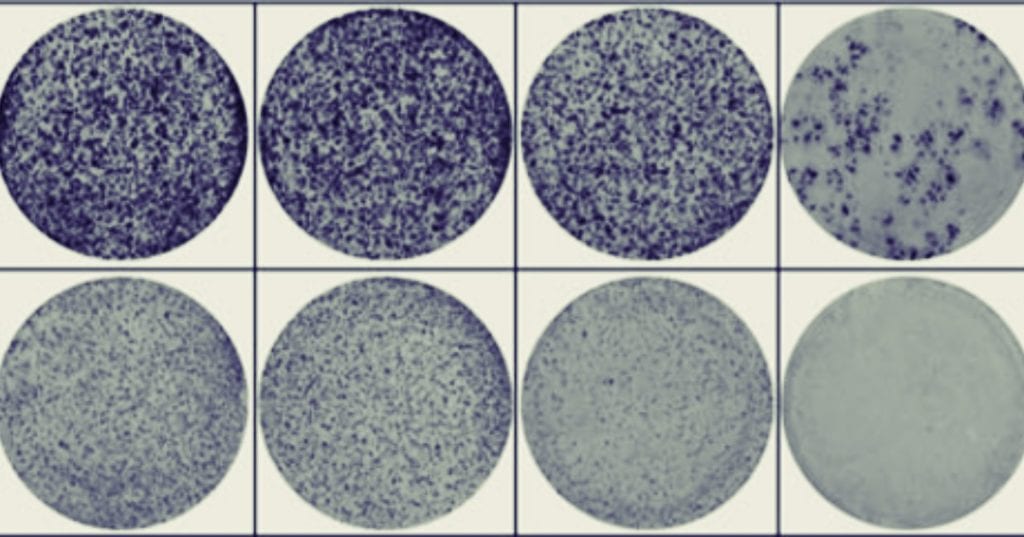
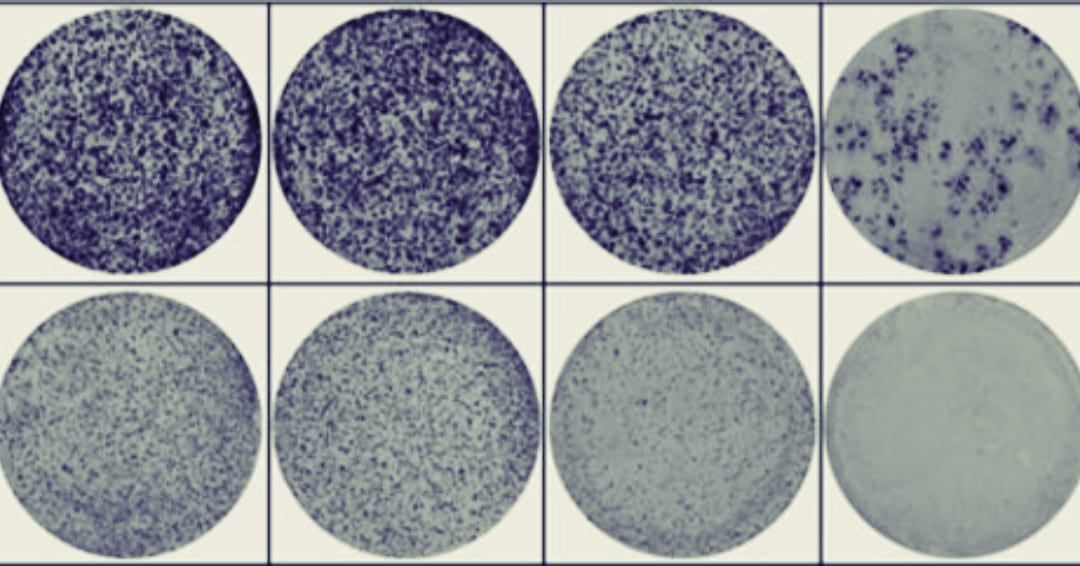
 Perhaps you’ve been there – looking at a colony formation assay (CFA) where there are hundreds (or thousands) of colonies of various sizes and wondering “how can I possibly count all of these colonies accurately”? The logistical problem becomes even more difficult when you realize you should only be counting colonies over 30μm and need to make sure not to recount previously counted colonies. The saddest part is even after you meticulously count this plate, statistically your count will be off by a coefficients of variation (CV) ranging from 8.1 to 40.0%1.
Perhaps you’ve been there – looking at a colony formation assay (CFA) where there are hundreds (or thousands) of colonies of various sizes and wondering “how can I possibly count all of these colonies accurately”? The logistical problem becomes even more difficult when you realize you should only be counting colonies over 30μm and need to make sure not to recount previously counted colonies. The saddest part is even after you meticulously count this plate, statistically your count will be off by a coefficients of variation (CV) ranging from 8.1 to 40.0%1.
In the paper “Activation of WNT/β-catenin signaling results in resistance to a dual PI3K/mTOR inhibitor in colorectal cancer cells harboring PIK3CA mutations”2 by Park et al. they have many high-density colony plates across their study (see Figure 1C, 5A, 5E and Supplementary Figure 2). To manage this, they forgo the typical situation where one researcher would manually count the colonies plate by plate and instead use an automated colony counter (GelCount).
Automated colony counters, particularly in a high-density situation like this, have a plethora of advantages over manual counting:
1) Automated colony counters can count and analyze four 6-well plates in under 15 minutes which would take a manual counter a great deal longer.
2) Automated counters don’t suffer from fatigue or inter/intra observer variation and will count every well the exact same way across an entire experiment.
3) Automated counters have the luxury to provide the user more information about colony growth dynamics like diameters, volumes and nearest neighbour distance.
4) Images can be stored in a bitmap format for presentations and publications. Alternatively, images can also be stored in a format to analyze and re-analyze colonies.
5) Growth dynamics from each and every colony can be exported to Excel and analyzed individually if needed. Again, this is not feasible via manual counts.
Park et al. are only one group of many who have used the automated colony counting methods to enhance and speed up research outputs. To read more articles that have used automated counting technology please follow this link here.
If you are interested to learn more or would be open to a demo of an automated colony counter, please feel free to reach out to sales@scintica.com
References:
- Standard Test Method for Automated Colony Forming Unit (CFU) Assays—Image Acquisition and Analysis Method for Enumerating and Characterizing Cells and Colonies in Culture (2012), ASTM International. DOI: 1520/F2944-12
- Park, Y. , Kim, H. , Cho, Y. , Min, D. , Cheon, S. , Lim, Y. J., Song, S. , Kim, S. J., Han, S. , Park, K. J. and Kim, T. (2019), Activation of WNT/β‐catenin signaling results in resistance to a dual PI3K/mTOR inhibitor in colorectal cancer cells harboring PIK3CA Int. J. Cancer, 144: 389-401. doi:10.1002/ijc.31662